-Search query
-Search result
Showing all 28 items for (author: ghosal & d)

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus
Method: helical / : Kreida S, Narita A, Johnson MD, Tocheva EI, Das A, Jensen GJ, Ghosal D

EMDB-28281: 
Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75
Method: subtomogram averaging / : Kaplan M, Ghosal D
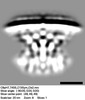
EMDB-28282: 
Subtomogram average of T4SS of Coxiella burnetii at pH 7
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D
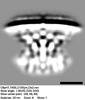
EMDB-28283: 
Subtomogram average of T4SS of Coxiella burnetii at pH7 with an inner membrane mask
Method: subtomogram averaging / : Kaplan M, Shepherd DC, Vankadari N, Kim KW, Larson CL, Przemyslaw D, Beare PA, Krzymowski E, Heinzen RA, Jensen GJ, Ghosal D

EMDB-14777: 
Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii
Method: helical / : Pakharukova N, Malmi H, Tuittila M, Paavilainen S, Ghosal D, Chang YW, Jensen GJ, Zavialov AV

PDB-7zl4: 
Cryo-EM structure of archaic chaperone-usher Csu pilus of Acinetobacter baumannii
Method: helical / : Pakharukova N, Malmi H, Tuittila M, Paavilainen S, Ghosal D, Chang YW, Jensen GJ, Zavialov AV

EMDB-20459: 
Anthrax toxin protective antigen channels bound to lethal factor
Method: single particle / : Hardenbrook NJ, Liu S

EMDB-20955: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S

EMDB-20957: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S

EMDB-20958: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S

PDB-6psn: 
Anthrax toxin protective antigen channels bound to lethal factor
Method: single particle / : Hardenbrook NJ, Liu S, Zhou K, Zhou ZH, Krantz BA

PDB-6uzb: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S, Zhou K, Zhou ZH, Krantz BA

PDB-6uzd: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S, Zhou K, Zhou ZH, Krantz BA

PDB-6uze: 
Anthrax toxin protective antigen channels bound to edema factor
Method: single particle / : Hardenbrook NJ, Liu S, Zhou K, Zhou ZH, Krantz BA

EMDB-20712: 
In vivo structure of the Legionella type II secretion system by electron cryotomography (aligning the IM-complex).
Method: subtomogram averaging / : Ghosal D, Jensen GJ

EMDB-20713: 
In vivo structure of the Legionella type II secretion system by electron cryotomography (aligning the OM-complex).
Method: subtomogram averaging / : Ghosal D, Jensen GJ

EMDB-0566: 
Subtomogram average of the Legionella pneumophila Dot/Icm type IV secretion system with DotF fused to a superfolder GFP (aligning the OM-complex)
Method: subtomogram averaging / : Ghosal D, Jeong KC, Chang YW, Vogel JP, Jensen GJ

EMDB-0464: 
Legionella pneumophila bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ

EMDB-0465: 
Pseudomonas aeruginosa bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ
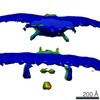
EMDB-0467: 
Shewanella oneidensis MR-1 bacterial flagellar motor
Method: subtomogram averaging / : Kaplan M, Ghosal D, Subramanian P, Oikonomou CO, Kjaer A, Pirbadian S, Ortega DR, Briegel A, El-Naggar MY, Jensen GJ

EMDB-7474: 
Subtomogram average of the cag type IV secretion system in Helicobacter pylori cells (aligning periplasmic parts)
Method: subtomogram averaging / : Chang YW, Shaffer C, Rettberg LA, Ghosal D, Jensen GJ

EMDB-7475: 
Subtomogram average of the cag type IV secretion system in Helicobacter pylori cells (aligning cytoplasmic parts)
Method: subtomogram averaging / : Chang YW, Shaffer C, Rettberg LA, Ghosal D, Jensen GJ

EMDB-8603: 
Sub-tomogram average of the archaellar motor in Thermococcus kodakaraensis
Method: subtomogram averaging / : Chang YW, Kjaer A, Briegel A, Oikonomou CM, Jensen GJ

EMDB-8566: 
In vivo structure of the Legionella pneumophila Dot/Icm type IV secretion system (aligning the outer membrane complex)
Method: subtomogram averaging / : Ghosal D, Chang YW, Jensen GJ

EMDB-8567: 
In vivo structure of the Legionella pneumophila Dot/Icm type IV secretion system (aligning the inner membrane complex)
Method: subtomogram averaging / : Ghosal D, Chang YW, Jensen GJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
